Pandas is an open source data analysis and manipulation tool, built on top of the Python programming language. Offers data structures and operations for manipulating numerical tables and time series.
Installation:
#checking version
import pandas as pd
print(pd.__version__)
Pandas Series
#series
vec = [0,12,99,54,89,10,28,30]
res = pd.Series(vec)
#print(res)
print(res[0])
print(res[2:])
print(res[:-4])
vec=[1,2,3,4]
own_index = pd.Series(vec,["I","II","III","IV"])
print(own_index)
#series with key values
random_values = {"I": 10, "II": 200, "III": 3000, "IV":40000}
res = pd.Series(random_values)
print(res)
Data Frames
#DataFrames
data = {
"cardio": [20, 26, 39, 41, 29],
"steps": [2700, 3400, 4520, 4700, 3700],
"weight": [65.0,64.5,63.0,62.0, 61.5]
}
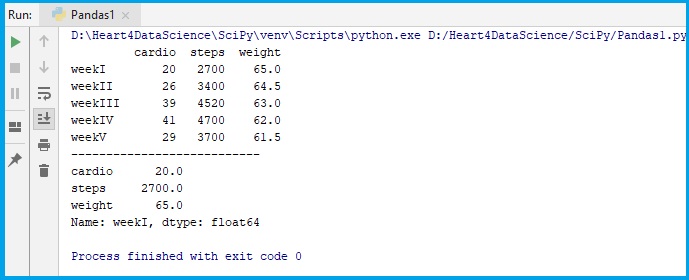
fit_table = pd.DataFrame(data, index = ["weekI", "weekII","weekIII","weekIV","weekV"])
print(fit_table)
#refers to one column only:
print(fit_table.loc["weekI"])
Stats
To see some results for various statistic tests we need te copy some data into PyCharm. I’ve created simple data file .csv :
import scipy.stats as sps
import pandas
FileData = pandas.read_csv('man_and_woman.csv', sep=',', na_values='.')
#T-test
T_test_result = sps.ttest_rel(FileData['Age'],FileData['IQ'])
T_test_result1 = sps.ttest_1samp(FileData['Age'] - FileData['IQ'],0)
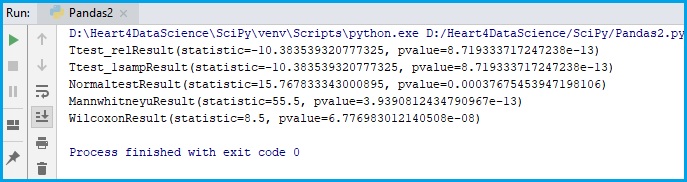
print(T_test_result)
print(T_test_result1)
#Normal test
normaltest_results = sps.normaltest(FileData['Age'])
print(normaltest_results)
# Mann-Whitney test
mw_results = sps.mannwhitneyu(FileData['Age'], FileData['IQ'])
print(mw_results)
# Wilcoxona test
wilcoxon_results = sps.wilcoxon(FileData['Age'], FileData['IQ'])
print(wilcoxon_results)
version with file and data processing in columns
example with data downloaded from Kaggle platform: Divorce/marriage dataset with birth dates
import scipy.stats as sps
import pandas
FileData = pandas.read_csv('divorces_2000-2015_translated.csv', sep=',', na_values='0')
# print(FileData.head())
# print(FileData.dtypes)
#T-test
T_test_result = sps.ttest_rel(FileData['Monthly_income_partner_woman_peso'],
FileData['Monthly_income_partner_man_peso'])
T_test_result1 = sps.ttest_1samp(FileData['Monthly_income_partner_woman_peso'],
FileData['Monthly_income_partner_man_peso'],0)
# print(T_test_result)
# print(T_test_result1)
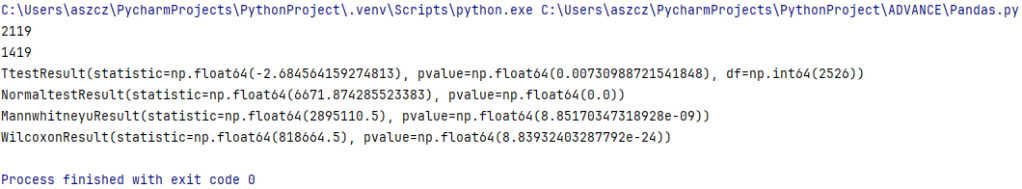
print(FileData['Monthly_income_partner_woman_peso'].isna().sum())
print(FileData['Monthly_income_partner_man_peso'].isna().sum())
# select columns from file
FileData_cols = ['Monthly_income_partner_woman_peso',
'Monthly_income_partner_man_peso']
FileData_clean = FileData[FileData_cols].dropna()
#delete rows with gaps in both columns at once
sps.ttest_rel(FileData_clean.iloc[:, 0], FileData_clean.iloc[:, 1])
#T-test
result = sps.ttest_rel(
FileData_clean['Monthly_income_partner_woman_peso'],
FileData_clean['Monthly_income_partner_man_peso']
)
print(result)
# Normal test
normaltest_results = sps.normaltest(FileData_clean['Monthly_income_partner_woman_peso'])
print(normaltest_results)
# Mann-Whitney test
mw_results = sps.mannwhitneyu(FileData_clean['Monthly_income_partner_woman_peso'],
FileData_clean['Monthly_income_partner_man_peso'])
print(mw_results)
# Wilcoxona test
wilcoxon_results = sps.wilcoxon(FileData_clean['Monthly_income_partner_woman_peso'],
FileData_clean['Monthly_income_partner_man_peso'])
print(wilcoxon_results)